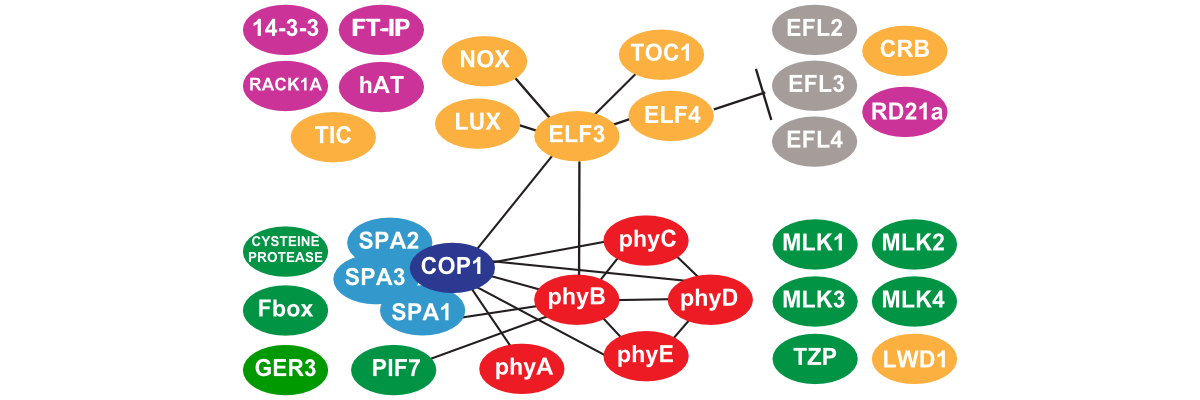
Deciphering the Circadian Interactome
Plants use an endogenous time keeping mechanism known as the circadian clock to regulate the timing of metabolism, physiology and development. However, the basic composition of the circadian system is unknown and how the clock is connected to other pathways in unclear. We combine affinity purification, mass spectrometry, and genetics to identify, dissect, and define the protein complexes within the clock.
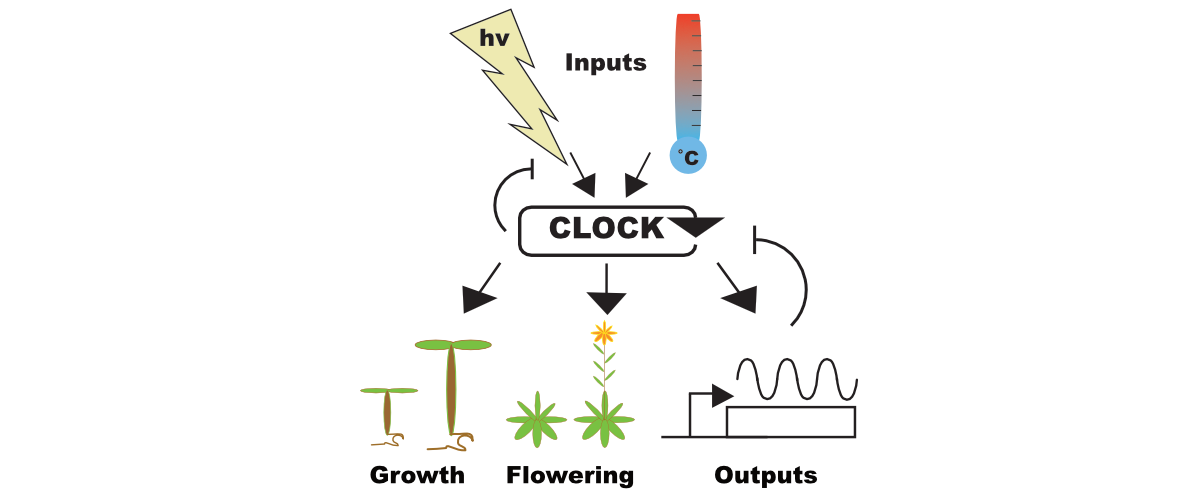
Circadian Regulation of Growth
The circadian clock regulates myriad processes in plants. We are particularly interested in how plants adapt their growth to seasonal variation in temperature and day length. We have identified new molecules that act as conduits between the circadian system and growth pathways. We are determining the function of these key proteins with the aim of improving crop production in response to a changing environment.
Tool development for Plants
In order to determine the molecular mechanisms that underlie key plant-specific processes, we are developing new tools and methods to explore protein function. This includes developing methods for rapid, facile purification of protein complexes and new methods for maintaining epitope-tagged proteins at endogenous levels and in their native context. We also develop inexpensive non-invasive imaging systems to monitor plant movement, bioluminescence and fluorescence during long-term time lapse experiments. We make the protocols, instructions, and scripts available and open-source so that we can leverage the community to improve and extend these when possible.
Publications
Preprint
Lee DY, Acosta-Gamboa L, Saleh L, Pathak S, Sawyer N, Morgan A, Meerdink S, Kuzio C, Calderon S, Sheng H, Kenney S, Zare A, Gehan M, Nusinow DA Remote sensing of endogenous pigmentation by inducible synthetic circuits in grasses bioRxiv 2025.06.20.660755; doi: https://doi.org/10.1101/2025.06.20.660755
–
Jawahir V, Adam S, Griffiths M, Wesley T, Phippen W, Heller N, Borphukan B, Sanguinet K, Nusinow DA Loss of PIF7 attenuates shade and elevated temperature responses throughout the lifecycle in Pennycress bioRxiv 2025.03.17.643809; doi: https://doi.org/10.1101/2025.03.17.643809
–
Kim SC, Nusinow DA, Wang X Identification of Arabidopsis phospholipase Ds involved in circadian clock alterations using CRISPR/Cas9-based multiplex editing bioRxiv 2024.01.09.574824; doi: https://doi.org/10.1101/2024.01.09.574824
–
2025
Cox KL Jr, Pardi SA, O'Connor L, Klebanovych A, Huss D, Nusinow DA, Meyers BC, Czymmek KJ ExPOSE: a comprehensive toolkit to perform expansion microscopy in plant protoplast systems Plant Journal (2025) Mar;121(5):e70049. doi: 10.1111/tpj.70049.
–
2024
Battle MW, Ewing SF, Dickson C, Obaje J, Edgeworth KN, Bindbeutel R, Kourounioti RA, Nusinow DA, Jones MA Manipulation of Photosensory and Circadian Signalling Restricts Phenotypic Plasticity and Dawn Anticipation in Arabidopsis Molecular Plant (2024) doi: 10.1016/j.molp.2024.07.007
–
Park SY, Qiu J, Wei S, Peterson FC, Beltrán J, Medina-Cucurella A, Vaidya A, Xing Z, Volkman BF, Nusinow DA, Whitehead TA, Wheeldon I, Cutler SR Orthogonal control of genetic circuits using a designed chemical-induced dimerization module with reprogrammable ligand binding specificity. Nature Chemical Biology (2024) 20, 103–110 https://doi.org/10.1038/s41589-023-01447-7
–
2023
Kim SC, Edgeworth KN, Nusinow DA, Wang X. Circadian clock factors regulate the first condensation reaction of fatty acid synthesis in Arabidopsis. Cell Reports (2023) Dec 26;42(12):113483. doi: 10.1016/j.celrep.2023.113483. PMID: 37995186x
–
Gao M, Lu Y, Geng F, Klose C, Staudt AM, Huang H, Nguyen D, Lan H, Mockler TC, Nusinow DA, Hiltbrunner A, Schäfer E, Wigge PA, and Jaeger KE CPhytochromes transmit photoperiod information via the evening complex in Brachypodium. Genome Biology (2023) Nov 7;24(1):256. doi: 10.1186/s13059-023-03082-w. PMID: 37936225; PMCID: PMC10631206
–
Sorkin ML, Tzeng SC, King S, Romanowski A, Kahle N, Bindbeutel R, Hiltbrunner A, Yanovsky MJ, Evans BS, Nusinow DA COLD REGULATED GENE 27 and 28 antagonize the transcriptional activity of the RVE8/LNK1/LNK2 circadian complex Plant Physiology (2023) 192-3, 2436–2456, https://doi.org/10.1093/plphys/kiad210
–
Sorkin ML, Markham KK, Zorich S, Menon A, Edgeworth KE, Ricono A, Bryant D, Bart R, Nusinow DA, Greenham K Assembly and operation of an imaging system for long-term monitoring of bioluminescent and fluorescent reporters in plants Plant Methods (2023) 19-19 doi:10.1186/s13007-023-00997-0
–
2022
Beltrán J, Steiner PJ, Bedewitz M, Wei S, Peterson FC, Li Z, Hughes BE, Hartley Z, Robertson NR, Medina-Cucurella AV, Baumer ZT, Leonard AC, Park SY, Volkman BF, Nusinow DA, Zhong W, Wheeldon I, Cutler SR, Whitehead TA. Rapid biosensor development using plant hormone receptors as reprogrammable scaffolds Nature Biotechnology (2022) Pub. June 20, 2022 doi: 10.1038/s41587-022-01364-5.
–
Steiner PJ, Swift SD, Bedewitz M, Wheeldon I, Cutler SR, Nusinow DA, Whitehead TA. A Closed Form Model for Molecular Ratchet-Type Chemically Induced Dimerization Modules Biochemistry (2022) Article ASAP doi: 10.1021/acs.biochem.2c00172
–
Zhang N, Pazouki L, Nguyen H, Jacobshagen S, Bigge BM, Xia M, Mattoon EM, Klebanovych A, Sorkin M, Nusinow DA, Avasthi P, Czymmek KJ, Zhang R.Comparative Phenotyping of Two Commonly Used Chlamydomonas reinhardtii Background Strains: CC-1690 (21gr) and CC-5325 (The CLiP Mutant Library Background) Plants. (2022) 11(5):585. doi: 10.3390/plants11050585
–
Sorkin ML, Nusinow DA Using Tandem Affinity Purification to Identify Circadian Clock Protein Complexes from Arabidopsis Methods Mol Biology (2022) 2398:189-203. doi: 10.1007/978-1-0716-1912-4_15. PubMed PMID: 34674177
–
2021
Griffiths M, Delory BM, Jawahir V, Wong KM, Bagnall GC, Dowd TG, Nusinow DA, Miller AJ, Topp CN Optimisation of root traits to provide enhanced ecosystem services in agricultural systems: A focus on cover crops Plant Cell Environment (2021) Dec 16;. doi: 10.1111/pce.14247. [Epub ahead of print] PubMed PMID: 34914117
–
Anderson CM, Mattoon EM, Zhang N, Becker E, McHargue W, Yang J, Patel D, Dautermann O, McAdam SAM, Tarin T, Pathak S, Avenson TJ, Berry J, Braud M, Niyogi KK, Wilson M, Nusinow DA, Vargas R, Czymmek KJ, Eveland AL, Zhang R High light and temperature reduce photosynthetic efficiency through different mechanisms in the C4 model Setaria viridis Communcation Biology (2021) Sep 16;4(1):1092. doi: 10.1038/s42003-021-02576-2. PubMed PMID: 34531541; PubMed Central PMCID: PMC8446033
–
Pardi SA, Nusinow DA Out of the Dark and Into the Light: A New View of Phytochrome Photobodies Front Plant Science 2021;12:732947. doi: 10.3389/fpls.2021.732947. eCollection 2021. Review. PubMed PMID: 34531891; PubMed Central PMCID: PMC8438518.
–
Sorkin Ml and Nusinow DA Time Will Tell: Intercellular Communication in the Plant Clock Trends in Plant Science (2021) doi: 10.1016/j.tplants.2020.12.009
–
Wilson ME, Tzeng SC, Augustin MM, Meyer M, Jiang X, Choi J, Bulter J, Evans B, Kutchan TM, Nusinow DA Quantitative Proteomics and Phosphoproteomics Support a Role for Mut9-Like Kinases in Multiple Metabolic and Signaling Pathways in Arabidopsis Molecular and Cellular Proteomics (2021) e100063 doi: 10.1016/j.mcpro.2021.100063
–
2020
Odipio J, Getu B, Chauhan RD, Alicai T, Bart R, Nusinow DA, Taylor NJ Transgenic overexpression of endogenous FLOWERING LOCUS T-like gene MeFT1 produces early flowering in cassava PLoS One (2020) 15(1): e0227199 doi: 10.1371/journal.pone.0227199
–
Chen WW, Takahashi N, Hirata Y, Ronald J, Porco S, Davis SJ, Nusinow DA, Kay SA, Mas P A mobile ELF4 delivers circadian temperature information from shoots to roots Nature Plants (2020) Apr;6(4):416-426. doi: 10.1038/s41477-020-0634-2
–
Nimmo HG, Laird J, Bindbeutel R, Nusinow DA The evening complex is central to the difference between the circadian clocks of Arabidopsis thaliana shoots and roots Physiologia Plantarum (2020) Apr 17;. doi: 10.1111/ppl.13108/p>
–
Mamidi S, Healy AS, Huang P, Grimwood J, Jenkins J, Barry K, Sreedasyam A, Shu S, Lovell JT, Feldman M, Wu J, Yu Y, Chen C, Johnson J, Sakakibara H, Kiba T, Sakurai T, Tavares R, Nusinow DA, Baxter I, Schmutz J, Brutnell TP, Kellogg EAA genome resource for green millet Setaria viridis enables discovery of agronomically valuable loci Nature Biotechnology (2020) 38, pages1203–1210 doi: 10.1038/s41587-020-0681-2>
–
2019
Huang H, McLoughlin KE, Sorkin ML, Burgie ES, Bindbeutel RK, Vierstra RD, Nusinow DA PCH1 regulates light, temperature, and circadian signaling as a structural component of phytochrome B-photobodies in Arabidopsis Proceedings of the National Academy of Sciences (2019) doi: 10.1073/pnas.1818217116
–
Kim SC, Nusinow DA, Sorkin ML, Pruneda-Paz J, Wang X Interaction and regulation between lipid mediator phosphatidic acid and circadian clock regulators in Arabidopsis. Plant Cell (2019) doi: 10.1105/tpc.18.00675
–
2018
Song YH, Kubota A, Kwon MS, Covington MF, Lee N, Taagen ER, Cintrón DL, Hwang DY, Reiko Akiyama R, Hodge SK, Huang H, Nguyen NH, Nusinow DA, Millar AJ, Shimizu KK, and Imaizumi T. Molecular basis of flowering under natural long-day conditions in Arabidopsis. Nature Plants (2018) doi: 10.1038/s41477-018-0253-3
–
Tovar JC, Hoyer JS, Lin A, Tiekling A, Callen ST, Castillo SE, Miller M, Monica Tessman M, Fahlgren N, Carrington JC, Nusinow DA, Gehan MA Raspberry Pi Powered Imaging for Plant Phenotyping Applications in Plant Sciences (2018) doi: 10.1002/aps3.1031
–
Khan MA, Castro-Guerrero NA, McInturf SA, Nguyen NT, Dame AN, Wang J, Bindbeutel RK, Joshi T, Jurisson SS, Nusinow DA, and Mendoza-Cozatl DG Changes in iron availability in Arabidopsis are rapidly sensed in the leaf vasculature and impaired sensing leads to opposite transcriptional programs in leaves and roots Plant, Cell and Environment (2018) doi: 10.1111/pce.13192
–
2017
Hughes ME, Abruzzi KC, Allada R, Anafi R, Arpat AB, Asher G, Baldi P, de Bekker C, Bell-Pedersen D, Blau J, Brown S, Ceriani MF, Chen Z, Chiu JC, Cox J, Crowell AM, DeBruyne JP, Dijk DJ, DiTacchio L, Doyle FJ, Duffield GE, Dunlap JC, Eckel-Mahan K, Esser KA, FitzGerald GA, Forger DB, Francey LJ, Fu YH, Gachon F, Gatfield D, de Goede P, Golden SS, Green C, Harer J, Harmer S, Haspel J, Hastings MH, Herzel H, Herzog ED, Hoffmann C, Hong C, Hughey JJ, Hurley JM, de la Iglesia HO, Johnson C, Kay SA, Koike N, Kornacker K, Kramer A, Lamia K, Leise T, Lewis SA, Li J, Li X, Liu AC, Loros JJ, Martino TA, Menet JS, Merrow M, Millar AJ, Mockler T, Naef F, Nagoshi E, Nitabach MN, Olmedo M, Nusinow DA, Ptáček LJ, Rand D, Reddy AB, Robles MS, Roenneberg T, Rosbash M, Ruben MD, Rund SSC, Sancar A, Sassone-Corsi P, Sehgal A, Sherrill-Mix S, Skene DJ, Storch KF, Takahashi JS, Ueda HR, Wang H, Weitz C, Westermark PO, Wijnen H, Xu Y, Wu G, Yoo SH, Young M, Zhang EE, Zielinski T, Hogenesch JB. Guidelines for Genome-Scale Analysis of Biological Rhythms Journal of Biological Rhythms (2017) doi: 10.1177/0748730417728663
–
Odipio J, Alicai T, Ingelbrecht I, Nusinow DA, Bart R, and Taylor N. Efficient CRISPR/CAS9 Genome Editing of Phytoene desaturase in Cassava Frontiers (2017): p1780. doi: 10.3389/fpls.2017.01780
–
Huang H, Gehan M, Huss SE, Alvarez S, Lizarraga C, Gruebbling EL, Gierer J, Naldrett MJ, Bindbeutel R, Evans BS, Mockler TC, Nusinow DA Cross-Species Complementation Reveals Conserved Functions For EARLY FLOWERING 3 Between Monocots And Dicots Plant Direct (2017) doi:10.1002/pld3.18
–
Lee CM, Adamchek C, Feke A, Nusinow DA, and Gendron JM Mapping Protein–Protein Interactions Using Affinity Purification and Mass Spectrometry Methods In Molecular Biology 1610, no. M110 (2017): doi:10.1007/978-1-4939-7003-2_15
–
2016
Huang H and Nusinow DA Tandem Purification of His6-3x FLAG Tagged Proteins for Mass Spectrometry from Arabidopsis. Bio-protocol (2016): doi:10.21769/BioProtoc.2060
–
Huang H and Nusinow DA Into the Evening: Complex Interactions in the Arabidopsis circadian clock. Trends in Genetics (2016): doi:10.1016/j.tig.2016.08.002
–
Mutka AM, Fentress SJ, Sher JW, Berry JC, Pretz C, Nusinow DA and Bart R Quantitative, image-based phenotyping methods provide insight into spatial and temporal dimensions of plant disease. Plant Physiology (2016): doi:10.1104/pp.16.00984
–
Huang H, Alvarez S, and Nusinow DA Data on the Identification of Protein Interactors with the Evening Complex and PCH1 in Arabidopsis Using Tandem Affinity Purification and Mass Spectrometry (TAP–MS) Data in Brief (2016): doi:10.1016/j.dib.2016.05.014
–
Huang H, Yoo CY, Bindbeutel RK, Goldsworthy J, Tielking A, Alvarez S, Naldrett M, Evans B, Chen M, and Nusinow DA. PCH1 integrates circadian and light-signaling pathways to control photoperiod-responsive growth in Arabidopsis. eLife. (2016) doi:10.7554/eLife.13292.
–
Huang H, Alvarez S, Bindbeutel RK, Shen Z, Naldrett MJ, Evans BS, Briggs SP, Hicks LM, Kay SA, and Nusinow DA. Identification of evening complex associated proteins in Arabidopsis by affinity purification and mass spectrometry. Molecular & Cellular Proteomics. (2016). doi:10.1074/mcp.M115.054064
–
2015
Kaiserli E, Paldi K, O’Donnell L, Batalov O, Pedmale UV, Nusinow DA, Kay SA, Chory J. Integration of light and photoperiodic signaling in transcriptional nuclear foci. Developmental Cell (2015) doi:10.1016/j,devcel.2015.10.008
–
Waadt R, Manalansan B, Rauniyar N, Munemasa S, Booker MA, Brandt B, Waadt C, Nusinow, DA, Kay SA, Kunz HH, Schumacher K, DeLong A, Yates JR, and Schroeder JI. Identification of OST1-Interacting Proteins Reveals Interactions with SnRK2-Type Protein Kinases and with PP2A-Type Protein Phosphatases That Function in ABA Responses. Plant Physiology (2015) doi:10.1104/pp.15.00575
–
2012
Chow BY, Helfer A, Nusinow DA, Kay SA. ELF3 recruitment to the PRR9 promoter requires other Evening Complex members in the Arabidopsis circadian clock. Plant Signaling and Behavior (2012) doi:10.4161/psb.18766
–
2011
Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farre EM, Kay SA. The ELF4-ELF3-LUX Complex Links the Circadian Clock to Diurnal Control of Hypocotyl Growth. Nature (2011) doi:10.1038/nature10182
–
Helfer A, Nusinow DA, Chow BY, Gehrke AR, Bulyk ML, Kay SA. LUX ARRHYTHMO Encodes a Nighttime Repressor of Circadian Gene Expression in the Arabidopsis Core Clock. Current Biology doi:10.1016/j.cub.2010.12.021

Dmitri A. Nusinow, Ph.D.
Principal Investigator
meter (at) danforthcenter.org
ORCID ID: 0000-0002-0497-1723

Dong-Yeon Lee, Ph.D.
Staff Scientist
Dlee (at) danforthcenter.org
ORCID ID:0000-0003-0218-1071

Vanessica Jawahir, Ph.D.
Postdoctoral Fellow
VJawahir (at) danforthcenter.org
ORCID ID: Coming Soon

Rebecca Bindbeutel, M.S.
Administrative Lab Manager
RBindbeutel (at) danforthcenter.org
ORCID ID: 0000-0001-7994-2073

Stefanie King
WH Danforth Fellow, Washington University Graduate
Sking (at) danforthcenter.org
ORCID ID: 0000-0002-8691-7084

Kristen Edgeworth
WH Danforth Fellow, Washington University Graduate Student
KEdgeworth (at) danforthcenter.org
ORCID ID: 0000-0002-4232-7330

Salma Adam
Laboratory technician
sadam (at) danforthcenter.org
ORCID ID: Coming Soon

Adele Ayers
Laboratory Assistant
AAyers (at) danforthcenter.org
ORCID ID: Coming Soon

Jennifer Keezle
Administrative Assistant
JKeezle (at) danforthcenter.org
ORCID ID: Coming Soon
Lab Alumni
2025
DanDan Zhang, Ph.D., RTP, North Carolina
2024
Sarah Pardi, Ph.D., UCLA, Los Angeles, CA
Madison Miederhoff, Mizzou, Columbia, MO, REU program
2023
Ashton Kish, Plastomics, St. Louis, MO
Maizy Wilcox, Gallaudet University, Washington D.C., REU program
Yuan Wang, Ph.D., KWS, Missouri
2022
Maria Sorkin, Ph.D., Ginko Bioworks, MassachusettsAbigail Senne, University of Colorado, Colorado Springs, CO, REU program
Evan Wang, John Burroughs High School, MO
2021
Maggie Wilson, Ph.D., Donald Danforth Plant Science Center, Missouri
Nathan Swyers, Ph.D., M6P Therapeutics, Missouri
Austin Morgan, (Graduate Student, University of Deleware)
Eileen Kosola, (University of Minnesota, Twin Cities, MN)
2020
Sunita Pathak M.S., (Graduate Student, Duke University, North Carolina)
2019
Rachel Tavares, (Graduate Student, U. Mass Amherst, Massachusetts)
Katherine Benza, (Graduate Student, Washington University in St. Louis, Missourt)
2018
Dr. He Huang, (Staff Scientist, Vanderbilt University, Tennessee)
Shara Chopra, Penn State University, NSF-REU
Abigail Hunt, Holt High School, Independent Intern
2017
Sarah Huss, CTC Genomics, Missouri
Mayla Ayers, Harris Stowe University, NSF–REU
Abigail Hunt, Holt High School, Independent Intern
2016
David Stroshein, Lawrence Technological University, NSF-REU (Graduate School, U of Arizona)
J'Laan Pittman, Washington University, St. Louis, uSTAR Program (Masters program, Chemistry, George Washington University)
2015
Dhruv Patel, Cornell, NSF-REU (Graduate School, UC Berkeley)
Hannah Lucas, U. of Wisconsin, Madison, NSF-REU (Graduate School, Washington U, St. Louis)
Ali Peet, St. Joseph’s Academy, Independent Intern
WIll Wolfe, John Burroughs High School, Independent Intern
2014
Jessica Goldsworthy, Michigan State University, NSF-REU (Pharm D. Program, Ferris State University)
Tom Liu, Ladue High School, STARS Intern
Allison Tielking, MICDS, Independent Intern
2013
Morgan Clark, Western Illinois University, NSF-REU (Chemist, Covance)
Elle Gruebbeling, Timberland High School, STARS Intern
2012
Savannah Est, Holt High School, Independent Intern
